5.5 The nucleus
The cell nucleus is the most conspicuous organelle in many eukaryotes, but it can be small and inconspicuous in fungi. This organelle has two major functions: storage of hereditary material and coordination of all cellular activities (metabolism, growth, with all the synthetic processes on which it depends, and cell division). It houses the eukaryotic cell’s chromosomes, and is the location for all molecular processes involving DNA: replication, recombination and transcription (copying DNA gene sequences into messenger-RNA). The nucleus is also where post-transcriptional steps in gene expression, such as RNA processing to remove introns, take place.
Genomic DNA molecules are extremely long. For example, yeast chromosome III, the first ever to be fully sequenced, comprises 3.15 × 105 bases and since adjacent bases are separated by 0.34 nm, the chromosomal molecule can be estimated to be about 107 μm long. Yeast cells vary, but are about 5 to 7 μm in size; so this one molecule is between 15 and 20 times longer than the cell that contains it. The molecule is only 2 nm in diameter, of course, but it’s quite evident that the nuclear DNA must be highly condensed to fit in the nucleus.
The compaction progresses at several levels. Fourteen turns of the DNA helix, a length of 146 bp (base pairs), complex with an octamer of histone proteins (two copies of each of histones H2A, H2B, H3 and H4,) into the nucleosome core particle and the nucleosomes then wind into a hierarchy of 10 nm fibres, 30 nm fibres, and chromosome loops, which end up as the chromatin of fully condensed chromosomes. In yeast there’s a length of about 45 bp of DNA between nucleosomes because the linker histone H1, which is involved in forming 30 nm fibres in animals and plants, is missing.
It’s important to appreciate that chromatin is not a static structure. Chromatin participates in the minute-by-minute working activities of the nucleus and there are many proteins that are recruited to modify and adapt chromatin to enable its DNA to be expressed. Histones are acetylated and deacetylated to modify chromatin structure. There are a number of distinct histone acetylase and deacetylase complexes that have gene-specific effects on transcription. DNA-binding activators and repressors can recruit these histone acetylases/deacetylases to particular gene promoters to locally modify chromatin structure. Other chromatin modifying complexes are responsible for creating larger regions of altered chromatin structure, associated with long-range effects on gene activity.
Most aspects of nuclear activity involve extremely large multiprotein complexes (often called molecular machines) and some of their components have chromatin-modifying activities. For example, the RNA transcription machinery and the molecular machine responsible for mRNA splicing are each comparable to ribosomes in terms of size and subunit complexity. As an aside: the Nobel Prize in Chemistry 2014 was awarded jointly to Jean-Pierre Sauvage, Sir J. Fraser Stoddart and Bernard L. Feringa ‘…for the design and synthesis of molecular machines…’ [see https://www.nobelprize.org/nobel_prizes/chemistry/laureates/2016/popular-chemistryprize2016.pdf].
The central dogma of molecular biology has traditionally been that RNA is a messenger molecule that exports the information coded into DNA out of the nucleus in order to code the synthesis of proteins in the cytoplasm: DNA → RNA → Protein. A ribonucleic acid polymerase (RNA polymerase or RNAP) is a multi-subunit enzyme that catalyses the process of transcription during which an RNA polymer is synthesised from a DNA template. Other RNAs well known to be involved in protein synthesis are transfer RNA (tRNA) and ribosomal RNA (rRNA). However, it is now clear that RNA serves a range of other functions. Some RNA molecules regulate gene expression, others act as enzymes and many have functions that are still unknown. These types of RNA are called non-coding or ncRNA, a category that includes microRNA (miRNA), small RNA (sRNA), interfering RNA (iRNA), small interfering RNA (siRNA) and antisense RNA.
Prokaryotes use the same RNAP to catalyse the polymerisation of coding as well as non-coding RNAs, eukaryotes have five distinct RNA polymerases.
- RNA polymerase I synthesises the major RNA molecules of the ribosome (which can account for nearly half of the RNA transcribed in a eukaryotic cell);
- RNA polymerase II produces all primary transcripts, which are the mRNA precursors, as well as small nuclear RNAs and micro RNAs;
- RNA polymerase III transcribes transfer RNAs, small ribosomal RNA and other small RNAs found in the nucleus and cytoplasm and which are necessary for normal functioning of the cell.
- RNA polymerases IV and V are found exclusively in plants; their function is essential for the formation of small interfering RNA and heterochromatin in the plant nucleus.
Messenger RNA transcription in fungi, particularly in the budding yeast Saccharomyces cerevisiae, is one of the main model systems for research on transcription in eukaryotes (Peñate & Chávez, 2014; Sesma & von der Haar, 2014), which requires a large set of proteins (called general transcription factors) to be assembled at the promoter before transcription can begin. These help the RNA polymerase to bind to the promoter, open up the double-stranded DNA and then switch RNA polymerase into elongation mode. Other proteins required for transcription initiation include activators binding to specific sequences to enhance attachment of polymerase; transcription mediators that interface the activators to the transcription factors, and other enzymes modifying chromatin structure to aid transcription by opening up the chromatin structure. As some of these proteins are themselves made up of more than one polypeptide, approximately 100 protein subunits must assemble at the promoter site to initiate transcription (Kornberg, 2007) (Fig. 1 shows a simplified overview). Once transcription is under way, most of the transcription factors detach from the polymerase complex.
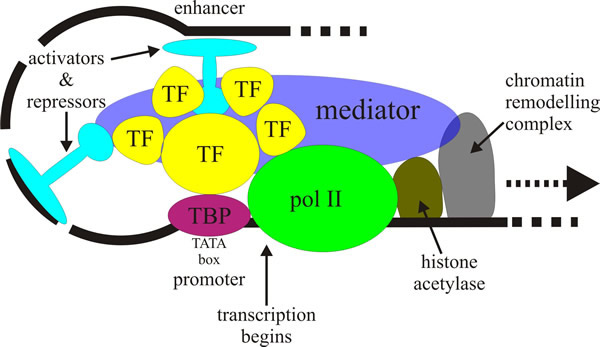 |
| Fig. 1. A simplified model of activation of
RNA polymerase II dependent transcription involving
assembly by gene-specific activator proteins of a pre-initiation complex
(PIC) that will eventually synthesise a specific messenger RNA. This
conversion requires structural changes in chromatin and assembly of general
transcription factors (TFs) and RNA polymerase II (pol II) at the gene’s
core promoter sequence, which surrounds the transcription start site of the
gene. A key event is the interaction of DNA-bound activators like the
TATA-box binding protein (TBP) with coactivators (shown here labelled TF,
but generally called TAFIIs [= TATA-box binding protein-associated
factor(s)]. TAFII250 is a scaffold for assembly of other TAFIIs with TBP
into a complex called TFIID. TBP first binds to the promoter and then
recruits TFIIB to join TFIID (and TFIIA if present). Before joining the PIC,
RNA polymerase II and TFIIF are bound together, being recruited by TFIIB.
Finally, RNA polymerase II recruits TFIIE, which further recruits TFIIH to
complete the PIC assembly. TFIID and TFIIB are the only components of the
preinitiation complex that can bind specifically to core promoter DNA.
Visit the Wikipedia entry at https://en.wikipedia.org/wiki/Eukaryotic_transcription, and/or the transcription animation at http://vcell.ndsu.nodak.edu/animations/transcription/index.htm for further explanation. |
The ‘gene specificity’ aspects of the binding events that contribute to assembly of the pre-initiation complex (PIC) seem to reside in the TATA binding protein (TBP, a subunit of TFIID) and TATA-box binding protein-associated factor(s) (TAFs) and co-activators that make up TFIID and TFIIB. Among the TAFs, TAFII250 seems to be particularly important as it regulates binding of TBP to DNA, binds core promoter initiator proteins, binds acetylated lysine residues in core histones, and possesses enzyme activities that modify histones and other transcription factors. These activities aid in positioning and stabilising TFIID at particular promoters, and alter chromatin structure at the promoter, creating a sharp bend in the promoter DNA, to allow assembly of transcription factors into the PIC. By so doing, TAFII250 converts signals for gene activation into effective transcription.
TFIIE joins the growing complex and recruits TFIIH which has protein kinase activity that phosphorylates RNA polymerase II within the C-terminal repeat domain, CTD. The CTD is an extension appended to the C terminus of the largest subunit of RNA polymerase II, which serves as a flexible binding scaffold for numerous nuclear factors; which factors bind being determined by the phosphorylation patterns on the CTD repeats.
TFIIH has DNA helicase activity to unwind the promoter DNA, and it recruits nucleotide-excision repair proteins. Subunits within TFIIH that have ATPase and helicase activity create negative superhelical tension in the DNA that causes approximately one turn of DNA to unwind and form the transcription bubble. The template strand of the transcription bubble engages with the RNA polymerase II active site and transcript-RNA synthesis begins. After synthesis of about 10 nucleotides of RNA, RNA polymerase II escapes the promoter region to transcribe the remainder of the gene.
In view of the significance of transcription factors in directing transcription to specific genes, you should not be surprised that in later discussions of cellular events we will frequently refer to the involvement of transcription regulators in control of so many features. Those regulators may be modifying chromatin structure and/or affecting the specificity and/or activity of transcription and/or RNA processing machinery. Many of the transcription regulators themselves work through multi-protein complexes (often called co-activators) and individual proteins can be components of completely different, and functionally distinct, co-activator complexes.
Transcriptional regulation and chromatin structure are intimately meshed together with the result that events occurring during initiation of transcription can regulate mRNA processing and so affect gene expression. The critical association occurs when the mRNA is first synthesised. Before mRNA can be used by ribosomes as a template for protein synthesis, it must be processed by the addition of a methylated cap (at its 5′end) and a polyadenylated (poly-A) tail (at its 3′end). Provision for the cap is made at the very start of transcription; provision for the poly-A tail is made at the end of transcription. After the transcription initiation complex has been established, the carboxyterminal end of RNA polymerase II is phosphorylated and this causes the enzyme to shift from the initiation mode to its elongation mode. Once transcription is under way, most of the transcription factors detach from the polymerase complex, their function to initiate transcription being complete.
Resources Box CLICK HERE to VISIT a YouTube animation describing TRANSCRIPTION
This is an animation belonging to the Virtual Cell Animation Collection produced by the Molecular and Cellular Biology Learning Center at North Dakota State University http://vcell.ndsu.nodak.edu/animations/home.htm |
The phosphorylated ‘tail’ of the polymerase interacts directly with proteins that carry out the RNA-capping, poly-A processing and splicing; in other words the transcription machinery recruits the different RNA-processing machines to the initial RNA transcript (which is usually called a pre-mRNA). These different machines share components; for example the cleavage polyadenylation specificity factor (CPSF) contains particular TAFs as subunits. This close ‘mechanical’ relationship, and the fact that newly synthesised RNA, the RNA polymerase and many mRNA processing factors are all close together in the nucleus, have strongly suggested that the different ‘machines’ are formed into an ‘mRNA factory’ that integrates synthesis and processing of mRNA.
As soon as the primary transcript, or pre-mRNA, is completed, RNA polymerase releases the already 5’-capped RNA molecules, and cleavage factors bind to specific nucleotide sequences in the molecule. The 3’ end of the pre-mRNA is then put into the correct configuration for cleavage and stabilising factors to be added to the complex. The Poly A polymerase now binds to the pre-mRNA and cleaves the 3’ end, allows the complex to dissociate, and synthesises the polyadenylated tail, by adding adenine nucleotide residues to the 3’ end. As the tail is synthesised, proteins bind to it, increasing the rate at which it is synthesised. When the polyadenylation process is completed the processed pre-mRNA (which still contains introns) is ready for the splicing process (Fig. 2).
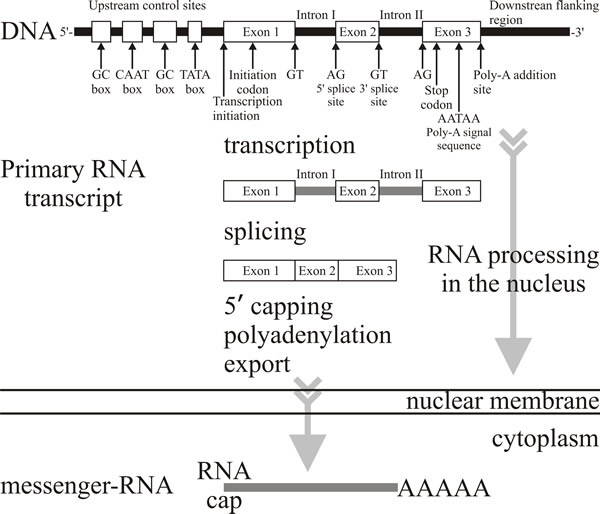 |
| Fig. 2. In eukaryotes the transcript of a protein-coding gene is called a primary transcript or pre-mRNA transcript and before it leaves the nucleus it is heavily modified by enzymes and ribozyme complexes. The 5′ end is capped with a modified guanine nucleotide; the 3′ end is polyadenylated with a sequence of up to 200 adenine nucleotides; finally the introns are removed by a process known as splicing. These processes are co-ordinated in time and space and actually occur as the pre-mRNA transcript is emerging from the RNA polymerase. Splicing involves linking the ends of two exons from a pre-mRNA transcript with high precision, discarding the intervening intron. The machinery involved uses 5 catalytic snRNA molecules (sn = small nuclear) and over 50 protein subunits. The assembly of snRNAs and proteins that perform the splicing is called the spliceosome. Splicing generates the fully-processed mRNA which is then exported to the cytoplasm to be translated (see Fig. 3 and the Resources Box animations, particularly the mRNA processing animation). |
Resources Box CLICK HERE to visit an animation describing mRNA PROCESSING
CLICK HERE to visit an animation describing
These animations belong to the Virtual Cell Animation Collection produced by the Molecular and Cellular Biology Learning Center at North Dakota State University http://vcell.ndsu.nodak.edu/animations/home.htm |
Intron and exon boundaries are defined by specific sequences in pre-mRNA which are recognised by a large array of snRNPs (small nuclear ribonucleoproteins) and other proteins that come together to form the reaction centres, the spliceosomes, on each spliced intron (Papasaikas & Valcárcel, 2016). The proteins are called SR proteins and have a common structure including one or more RNA-binding domains and a domain rich in arginine–serine dipeptides that function in the protein-protein interactions involved in splicing, transport and localisation of these proteins. Splicing involves linking the ends of two exons from a pre-mRNA transcript with high precision, and discarding the intervening intron. The machinery involved uses an assembly of five catalytic snRNA molecules and over 50 protein subunits called the spliceosome. The process involves changes in protein conformation that effectively loop out the intron (into a branched loop structure called a lariat) so that the enzymes in the spliceosome can cleave out the intron and join the ends of adjacent exons (Fig. 3). Splicing generates the fully-processed mRNA which is then exported to the cytoplasm for translation.
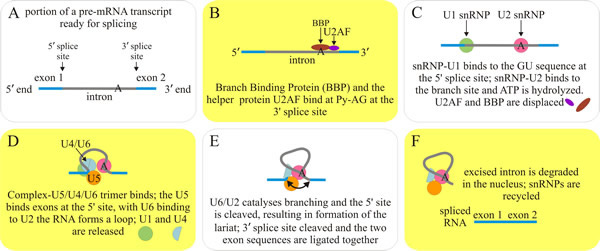 |
Fig. 3. The splicing process conducted by the major spliceosome protein-RNA assembly, which splices introns containing GU at the 5' splice site and AG at the 3' splice site and accounts for more than 99% of splicing activity in eukaryotes. The first step involves two complexes that bind at the Py-AG at the 3' splice site: Branch Binding Protein (BBP also called SF1 (splicing factor 1) in mammalian systems) and the helper protein U2AF. The RNA is looped, and three other protein-RNA complexes bind. This final complex undergoes a conformation change and the intron is cleaved at the 5′ GU sequence and forms a lariat at the A branch site. The 3′ end of the intron is next cleaved at the AG sequence, and the two exons are ligated together. As the spliced mRNA is released from the spliceosome, the intron debranches, and is then degraded. Visit the Wikipedia entry at http://en.wikipedia.org/wiki/RNA_splicing, and/or visit the mRNA splicing animation for further explanation. |
Throughout their time in the nucleus, RNA transcripts are associated with heterogeneous nuclear ribonucleoproteins (hnRNPs), an abundant family of RNA-binding proteins. These proteins are involved in almost every aspect of pre-mRNA processing as well as in mRNA transport and in translation. Evidently, the fate and function of the transcription product are intimately dependent on RNP complexes composed of the transcribed RNA and a wide variety of proteins.
Updated July, 2019