8.3 Mating type switching in budding yeast
Homothallic yeast cells can, remarkably, switch their mating type as often as every generation by a highly regulated, site-specific homologous recombination event that replaces one MAT allele with a different DNA sequence that encodes the opposite MAT allele. This replacement process involves the participation of two intact but unexpressed copies of mating-type information at the heterochromatic loci, HMLα and HMRa, which are located at opposite ends (Left and Right, respectively) of the same chromosome that encodes MAT.
Saccharomyces cerevisiae is heterothallic but a clone of haploid cells of the same mating type frequently sporulates, and there will be equal numbers of a and α cells amongst the progeny. This results from mating type switching controlled by the gene HO (HOmothallic) that exists in two allelic forms (dominant HO and recessive ho), and encodes an endonuclease. On either side of the MAT locus, and on the same chromosome, there are silent storage loci for each mating type, called HML and HMR. The HO/ho endonuclease creates a double-strand break at the MAT locus that initiates switching of information, by an homologous recombination event between the two parts of the same chromosome, at the MAT locus with that at either HML or HMR (Fig. 4).
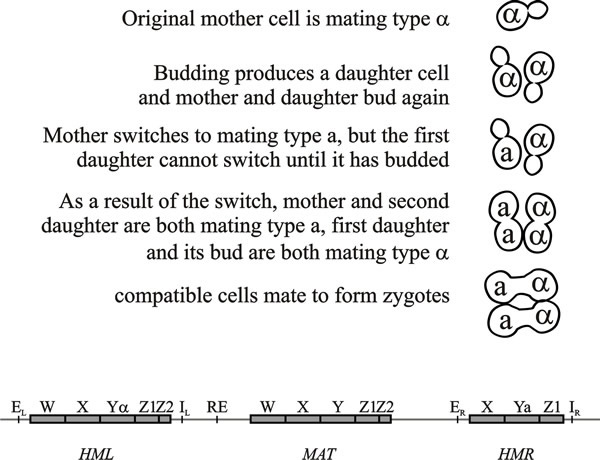 |
| Fig. 4. Top: pattern of mating type switching in Saccharomyces cerevisiae showing the consequences of a mating type switch in one mother cell. Bottom: the three loci involved in mating type switching, HML, MAT and HMR, are located on the same chromosome (not drawn to scale). HML is about 180 kb from MAT, and HMR about 120 kb from MAT; the centromere is located between RE and the MAT locus. A double strand break at the MAT locus, caused by the HO endonuclease, initiates a gene conversion event that replaces the Y region of the MAT locus with Y sequences from one of the storage loci. HML and HMR contain complete copies of the mating type genes but are not expressed because they have a repressed heterochromatin structure imposed by the E and I silencer sequences. HML shares more of the MAT sequences (W, X, Z1 and Z2) than does HMR. RE is a recombination enhancer that controls preferential recombination between MATa and HML, or between MATa and HMR. Modified from Chapter 2 in Moore & Novak Frazer, 2002. |
Again, MAT heterozygosity plays a key role in controlling the switching of mating-type genes. Switching occurs in homothallic strains expressing the HO endonuclease gene, HO, which are able to switch from MATa to MATα and vice versa, but once cells of opposite mating type conjugate to form a diploid, HO expression is repressed by the a1-α2 repressor. The Mata1, Matα1, and Matα2 transcription regulators that organise haploid cell specificity are quite rapidly turned over, being degraded by ubiquitin-mediated proteolysis by the proteasome; in contrast, the a1-α2 corepressor is much more stable.
Since yeasts can live in very small habitats, like flower nectaries and surfaces of individual fruits, yeast populations can be very isolated from one another in nature, so the rare mating type switching will give isolated populations the opportunity to undergo sexual reproduction; this is presumably its selective advantage. Mating type switching occurs about once in 105 divisions in cultures carrying allele ho, whereas in strains carrying HO the switch occurs at every cell division. However, there is an asymmetry in the cell division in that a new daughter bud is not able to switch mating types until it has itself budded. In S. cerevisiae, this is achieved by actively transporting the mRNA of a gene called Ash1 into the budding daughter cell. This mRNA encodes an inhibitor of the HO-endonuclease. Consequently, immediately after each division switching by the daughter is inhibited and only the mother cell is switchable. This means that even if there is only one cell to start with, a single division cycle will produce two cells of opposite mating type.
Saccharomyces cerevisiae has evolved an elaborate set of mechanisms to enable cells to switch their mating types. MAT switching depends on four phenomena:
- Presence of two unexpressed (silenced or cryptic) copies of mating-type sequences that act as donors during MAT switching. This implies that there is a mechanism which influences chromatin structure to maintain it in a silent configuration. The mechanism involves silencer sequences surrounding HML and HMR that interact, directly or indirectly, with several protein factors to repress the transcription of these genes. Among these are four Silent Information Regulator (Sir) proteins, a set of silencer binding proteins, histone proteins, the Rap1 protein, as well as several chromatin modifiers. Together, these create short regions (about 3 kb) of heterochromatin, in which the DNA sequences of HML and HMR are in a highly ordered nucleosome structure, known as heterochromatin, which is not transcribed by either RNA polymerase II or RNA polymerase III, and is resistant to cleavage by several endonucleases, including the HO endonuclease.
- Programmed creation of a site-specific double-strand break at MAT that results in the replacement of Ya or Yα sequences. HO is a site-specific endonuclease that recognises a 24-bp sequence that spans the MAT-Y/Z border. Haploid yeast has three possible targets for HO: the MAT locus, HMLα, and HMRa, but only the MAT locus is accessible under normal conditions because of gene silencing of HML and HMR. Normally the HO gene is tightly regulated and is expressed only in haploid mother cells and only at the G1 stage of the cell cycle. The result in mother cells is a single programmed double-strand break at the MAT locus prior to the initiation of DNA replication.
- A cell lineage rule ensuring that any cell that has previously divided once can switch MAT, while new daughter cells cannot (so only half of the cells in a population switch at any one time). The way this works is that a germinating haploid spore grows, produces a bud, and divides without changing mating type. Then, in the next cell division cycle, the older mother cell and its second daughter change mating type while the first daughter buds and divides without changing. This ensures that there will be cells of both mating types in close proximity; they readily conjugate, forming MATa/MATα diploids in which the HO endonuclease gene is turned off so that further mating-type switching is repressed. Control of this lineage pattern depends on expression of the HO gene being restricted to mother cells that have divided by the Swi5 transcription factor, which is localised to the mother cell nucleus and not that of her daughter. Lack of Swi5 expression in daughters is caused by the Ash1 repressor protein that is found only in the daughter cell. Ash1 may directly repress SWI5 gene transcription, thus restricting HO expression to the mother cell in the next G1 stage of the cell cycle. Ash1 mRNA is localised to the daughter prior to cell division by a myosin-like protein, actin, cargo binding proteins, translation repressors and a nuclear localising protein. Since Ash1 mRNA localisation was first discovered, many other mRNAs have been found to show similar localisation in Saccharomyces.
- A mechanism regulates the selective use of the two donors (called donor preference); study of which has yielded much of what we know about double strand break-induced recombination during eukaryote mitosis. Switching one mating type to the other involves the replacement at the MAT locus of Ya or Yα by a gene conversion induced by the site-specific double-strand break at MAT caused by HO endonuclease; it is a DNA-damage repair process. The process is highly directional, in that the sequences at MAT are replaced by copying new sequences from either HMLα or HMRa (whichever is the alternate to the resident sequence), while the two donor gene loci remain unchanged. HO endonuclease cannot cleave its recognition sequence at either HML or HMR, as these sites are occluded by nucleosomes in silenced DNA and this prevents crossing over. Any DNA-single strand exchanges (Holliday junctions) that might otherwise become crossovers are removed by two helicase enzymes, Sgs1 and Mph1, working with their partner proteins. Thus the (resident) MAT locus is cleaved and becomes the recipient in this gene conversion process. Overall, following HO cleavage of MATa, the end of the broken DNA molecules is excised in a 5′ to 3′ direction, creating a 3′-ended single-strand DNA tail on which assembles a filament of the Rad51 recombinase protein. Rad51 is essential for repairing damaged DNA and is highly conserved in most eukaryotes; this family of proteins interact with several other single-strand DNA-binding proteins to form a helical nucleoprotein filament on the DNA. This protein-DNA complex engages in a search for a homologous DNA sequence (since we started with MATa, in this case it would search for HMLα) to effect the repair. This search culminates in strand exchange in which the single stranded DNA base pairs with the complementary sequence of the donor, creating a displacement loop (D-loop). The 3′ end of the invading strand is then used as a primer to initiate copying of one strand of the donor locus, and the newly copied strand is displaced until it can anneal with homologous sequences on the opposite end of the double strand break. The 3′-ended nonhomologous tail is clipped off and the new 3′ end is used to prime a second strand of DNA synthesis, completing the replacement of MATa with MATα.
These important mechanisms (reviewed by Haber, 2012) are more examples of how research on fungi has informed our fundamental knowledge of the molecular genetics of eukaryotes.
The switching system of the related yeast, Kluyveromyces lactis, is similar to that of Saccharomyces cerevisiae to some extent, and includes sequences recognisably similar to MATa1, MATα1, and MATα2, but the shared flanking sequences are not closely related to the W, X, or Z1/Z2 sequences of Saccharomyces cerevisiae and there is no functional HO gene. While the Kluyveromyces lactis HMR has a1 and a2; HML includes a novel gene, α3, in addition to α1 and α2. Both HML and HMR are silenced by a mechanism dependent on Sir-proteins as in S. cerevisiae. Switching is dependent on a protein (Mts1) that is the homologue of the S. cerevisiae RME1 repressor; but in K. lactis it is required to activate switching and is turned off in MATa/MATα cells by an a1-α2 repressor. The major difference between the two yeasts, at least for MATα to MATa switching, is that it is the α3 gene, which is a transposable element that can excise from the DNA as a circle and somehow catalyse switching (Barsoum et al. 2010). MATa to MATα switching seems to be under the control of a different transposable element (Haber, 2012).
Mating type switching also occurs in the more distantly related fission yeast Schizosaccharomyces pombe, but the switching system differs in almost every detail from Saccharomyces cerevisiae. Mating-type switching in Schizosaccharomyces pombe involves replacing genetic information at the expressed mat1 locus with sequences copied from one of two silent donor loci, mat2-P or mat3-M, which are close together and located only a short distance away from mat1 on the same chromosome arm in a 20-kb length of heterochromatin. There is no HO-like enzyme; instead, a persistent single-strand break is created in the DNA at mat1, which is converted to a double strand break when cells enter S phase. Only one of the two daughter cells can switch. Donor selection is dictated by cell type: mat2 is the preferred donor in M cells, and mat3 is the preferred donor in P cells. Donor preference involves major changes in chromatin modification and structure and the silencing system is different from that in Saccharomyces cerevisiae (Jia et al. 2004).
Yeasts in a third clade of the Saccharomycotina, the methylotrophs, have a simpler two-locus switching system based on reversible inversion of a section of chromosome with MATa genes at one end and MATα genes at the other end. In Hansenula polymorpha the invertible region, which is 19 kb long, lies beside a centromere so that, depending on the orientation, either MATa or MATα is silenced by centromeric heterochromatin. In Pichia pastoris, the orientation of a 138 kb invertible region puts either MATa or MATα beside a telomere where heterochromatin silences MATa2 or MATα2. Both species are homothallic, and inversion of their MAT regions can be induced by crossing two strains of the same mating type. The three-locus Saccharomyces cerevisiae system may have been derived from mating-type switching by chromosomal inversion as seen in methylotrophic yeasts; the increased complexity of the S. cerevisiae switching apparatus, with three loci, donor bias, and cell lineage tracking, resulting from continuous evolutionary selection to increase sporulation ability in young colonies (Hanson et al. 2014).
Mating type switching, which is often referred to as stochastic (meaning randomised) mating type determination, has evolved independently in a number of organisms other than the yeasts; for example, in the ciliates, Tetrahymena thermophila, Paramecium spp., and Euplotes crassus. There is also evidence for some degree of randomised mating type identity during vegetative growth in the green algae Chlamydomonas monoica and Closterium ehrenbergii, and the dinoflagellate Gymnodinium catenatum, although the switching mechanism in these species is not known. Mating types in filamentous fungi tend to be far more stable. Oddly enough, switching does not occur in any of the best-studied organisms like Neurospora, Aspergillus, or Podospora, but has been claimed in Chromocrea spinulosa, Sclerotinia trifoliorum, Glomerella cingulata and Ophiostoma ulmi (Tsui et al., 2013), and the basidiomycete Agrocybe aegerita (Hadjivasiliou et al. 2016).
Updated July, 2019
