8.2 Mating in budding yeast
The life cycle of the yeast Saccharomyces cerevisiae features an
alternation of a haploid phase with a true diploid phase (Fig. 1), and in this
respect differs from filamentous Ascomycota in which the growth phase after
anastomosis is a heterokaryon.
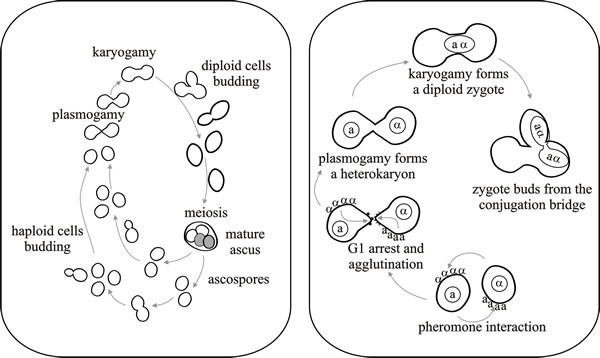 |
| Fig. 1. Life cycle (left hand panel) and mating process of the yeast Saccharomyces cerevisiae. Yeast can reproduce asexually by budding. Haploid cells of different mating types fuse to form dumbbell-shaped zygotes, which can themselves bud to establish a diploid clone. Well-nourished diploid cells, which are exposed to starvation conditions, enter meiosis, forming a 4-spored ascus. Ascospores germinate by budding. In the laboratory, ascospores can be separated to form haploid clones but in nature ascospores usually mate immediately, so the haploid phase is greatly reduced. The right hand panel depicts pheromone interaction, agglutination and the mating process in a little more detail. Modified from Chapter 2 in Moore & Novak Frazer, 2002. |
There are two mating types: haploid yeast cells may be of mating type a, or α. Karyogamy (nuclear fusion) follows the fusion of cells of opposite mating type and then the next daughter cell that is budded off contains a diploid nucleus. Most natural yeast populations are diploid because the haploid meiotic products mate while they are still close together immediately after the meiosis is completed. Diploid cells reproduce vegetatively by mitosis and budding until particular environmental conditions (deficiency in nitrogen and carbohydrate, but well aerated and with acetate or other carbon sources which favour the glyoxylate shunt) induce sporulation. When that happens, the entire cell becomes an ascus mother cell; meiosis occurs and haploid ascospores are produced. Ascospore germination re-establishes the haploid phase, which is itself maintained by mitosis and budding if the spores are separated from one another (by laboratory experiment, or by some disturbance in nature) to prevent immediate mating.
Mating type factors of yeast specify peptide hormones; these are called pheromones (the term originally applied to mate-attracting hormones of insects and mammals) and there are both pheromone α- (alpha-) and a-factors (Fig. 2), and corresponding receptors specific for each pheromone.
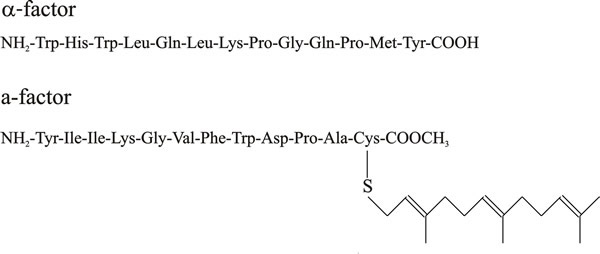 |
| Fig. 2. Simplified chemical structures of yeast pheromones. |
Pheromones organise the mating process; they have no effect on cells of the same mating type or on diploids but their binding to pheromone receptors on the surface of cells of opposite mating type (Fig. 1) act through GTP binding proteins to alter metabolism and:
- cause recipient cells to produce an agglutinin that enables cells of opposite mating type to adhere;
- stop growth in the G1 stage of the cell cycle;
- change wall structure to alter the shape of the cell into elongated projections;
- fusion eventually occurs between the projections.
In Saccharomyces cerevisiae a relatively small region of the genome, less than a thousand base pairs, codes for a complex genetic locus called MAT. The MAT locus lies in the middle of the right arm of chromosome III, about 100 kb from both the centromere and the telomere. The two mating-type alleles, MATα and MATa, differ by about 700 bp of sequences (designated Yα and Ya respectively) that contain the promoters and most of the open reading frames for proteins that regulate many aspects of the cell’s sexual activity (for review, see Haber, 2007). The MAT locus is divided into five regions (called W, X, Y, Z1, and Z2) on the basis of sequences that are shared between MAT and the two unexpressed (cryptic) copies of mating-type sequences that serve as donors during the recombinational process that allows a MATa cell to switch to MATα or vice versa (see Section 8.3 below).
The MATa locus encodes a1 and a2 polypeptides, the messengers for which are transcribed in opposite directions (Fig. 3), and MATα encodes polypeptides α1 and α2, all of which are transcription factors responsible for orchestrating both haploid cell type specificity (a or alpha) and the fate of the diploid zygote (a/alpha). Two are homeodomain proteins of the HD1 and HD2 class that form a heterodimer, a1/α2, so heterozygosity at MAT is a sign of diploidy and eligibility to sporulate; even partial diploids carrying MATa/MATα will attempt to sporulate. Analysis of the MATa1, MATα1, and MATα2 genes provided one of the earliest models of cell-type specification by transcriptional activators and repressors. MATα encodes two proteins, MATα1 and MATα2. MATα1, in conjunction with a constitutively expressed protein called Mcm1, activates a set of α-specific genes that includes genes encoding the mating pheromone (α-factor), and Ste2, a trans-membrane receptor of the opposite mating pheromone (a-factor). As mentioned above, these mating pheromones stop the cell cycle in the G1 stage, thereby promoting conjugation and ensuring that the zygote contains two unreplicated nuclei.
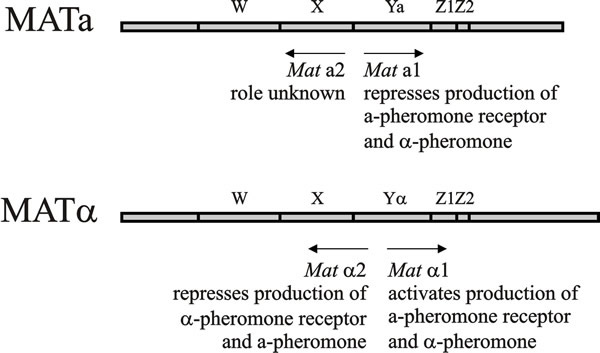 |
| Fig. 3. Functional domains in mating type factors of Saccharomyces cerevisiae. Region Y is the location of the mating type idiomorphs, which have very little homology with each other. Ya is 642 bp long, Yα is 747 bp long. Regions W, X, and Z1 and Z2 are homologous terminal regions. The arrows indicate direction of transcription and the legends beneath the arrows indicate functions of the gene products. In S. cerevisiae of mating type a, a general transcription activator is responsible for production of a-pheromone and the membrane-bound α-pheromone receptor. In a/α diploids, the MATa1/MATα2 heterodimer protein activates meiotic and sporulation functions, and represses haploid functions (turning off α-specific functions by repressing MATα1, a-specific functions being repressed by MATα2 alone). Modified from Chapter 2 in Moore & Novak Frazer, 2002. |
MATα2 encodes a homeodomain protein that acts with the constitutive Mcm1 to form a repressor of a-specific genes including those that produce a-factor pheromone (MFa1and MFa2) and the Ste3 transmembrane receptor protein that detects the presence of α-factor in the medium. Repression also requires the action of two other proteins, Tup1 and Ssn6. The MATa locus has two open reading frames MATa1 and MATa2 that are divergently transcribed (that is, they are transcribed in opposite directions).
Although MATa2 is well conserved evolutionarily, it has yet to be assigned a biological function. MATa2 and MATα2 share part of the same protein sequence and the evolutionary preservation of MATα2 may account for conservation of MATa2, despite its lack of apparent function. If MAT is entirely deleted, haploid cells mate identically to MATa cells (that is, the MATa phenotype is the default phenotype), because a-specific genes are constitutively expressed in the absence of Matα2 and α-specific genes are not transcribed in the absence of Matα1. Although MATa is not required for a-mating, the MATa1 gene is required, to work with MATα2 to repress haploid-specific gene expression, and this includes a gene called RME1, which is itself a repressor of meiosis and sporulation (RME1 means Repressor of MEiosis-1). Repression of this repressor permits expression of diploid-specific attributes (Haber, 2012).
Updated July, 2019
