White rot fungi
White rot fungi are basidiomycetes that are capable of degrading the lignin component of lignocellulose substrates (see Lignin structure for a short explanation). There are other fungi capable of digesting lignocellulose, such as brown rot fungi, but they do not produce the same ligninolytic enzymes and tend to concentrate their activities on the cellulose component. White rot fungi are so called because the degradation process results in a bleaching of the wood substrate (it's the polyphenolic lignin that provides most of the colour to native timber). White rot fungi are robust organisms that have a high tolerance to toxic environments, making them ideal to use for bioremedial purposes. They can also withstand high temperatures and a wide range of pH, further enhancing their hardy capabilities.
Oxidative lignin breakdown depends on a panel of enzymes including:
- lignin peroxidase, (a haem (Fe)-containing protein) which catalyses H2O2-dependent oxidation of lignin;
- manganese peroxidase, which also catalyses H2O2-dependent oxidation of lignin;
- laccase (a copper-containing protein) which catalyses demethylation of lignin components.
These are the key enzymes, and are described in a little more detail below. In addition, glyoxal oxidase, an extracellular peroxide-generating enzyme, and veratryl alcohol, which is a degradation product of lignin, also have important functions in lignin break down.
The process of catabolic lignin degradation involves:
- cleavage of ether bonds between monomers;
- oxidative cleavage of the propane side chain;
- demethylation;
- benzene ring cleavage to ketoadipic acid which is fed into the tricarboxylic acid cycle as a fatty acid.
Most research has been concentrated on white-rot basidiomycete fungi, such as Phanerochaete chrysosporium (Sporotrichum pulverulentum), which is able to mineralise lignin completely to CO2 and water. The lignin-degradative system of Phanerochaete chrysosporium appears after cessation of primary growth (that is, it is an aspect of the secondary metabolism of the organism) and can be induced by nitrogen starvation. Genomes of white-rot fungi feature families of genes encoding laccases, lignin peroxidases and manganese peroxidases. The multiplicity of these extracellular enzymes may be a result of their having multiple roles in fungal physiology (laccases, for example, contribute to plant pathogenesis, sporulation and pigment formation), but could also be a response to the diversity of the lignin substrate.
Lignin peroxidase (ligninase) is the key lignin-degrading enzyme of white-rot fungi. The Phanerochaete chrysosporium family of lignin peroxidases comprises extracellular glycosylated heme proteins, which are secreted in response to nitrogen limitation. When the fungus is grown in low-nitrogen medium there is an increase in H2O2 production by cell extracts which correlates with the appearance of ligninolytic activity; experimental destruction of H2O2 by adding the enzyme catalase strongly inhibits lignin breakdown. Activated oxygen derived from H2O2 is involved in degrading lignin, but is held in the active site of a specific extracellular enzyme, the lignin peroxidase. Lignin peroxidases are strongly oxidative; the enzyme is activated by itself being oxidised by H2O2, the initial step involving oxidation by one electron to produce an unstable intermediate which is then able to catalyse oxidation of phenols, aromatic amines, aromatic ethers and polycyclic aromatic hydrocarbons. Veratryl alcohol is a secondary metabolite that stimulates lignin degradation by recycling the lignin peroxidase and protecting the enzyme against inactivation by H2O2.
There can be as many as 15 lignin peroxidase isozymes, ranging in molecular mass from 38,000 to 43,000, the spectrum of isozymes produced depending on culture conditions and strains employed. However, ten lignin peroxidase genes, with a conserved sequence, have been identified in Phanerochaete chrysosporium and mapped into three different linkage groups. Lignin peroxidases are not found in all white-rot fungi, though. In particular, they seem to be absent from Ceriporiopsis subvermispora, a white-rot basidiomycete that is widely studied for its potential use in the pulp and paper industry (‘biopulping’) and in which manganese peroxidases are responsible for ligninolysis.
Manganese-dependent peroxidases are another family of extracellular glycosylated haem proteins, which are produced by most white-rot fungi. Like lignin peroxidase, the manganese peroxidases also require H2O2 to function, but the mechanism is very different. The manganese peroxidase system creates low-molecular-weight oxidising agents that diffuse into the lignin substrate and are able to oxidise phenolics residues in the lignin some distance from the enzyme. Some of these oxidising agents may be peroxidated lipids, but the chief one, from which the enzyme gets its name, is the metal ion manganese; the enzyme uses H2O2 to oxidise extracellular Mn(II) to Mn(III) and this becomes the diffusible oxidant that can degrade lignin at a distance.
The H2O2 which is required by peroxidases is probably generated by glyoxal oxidase, an extracellular enzyme that transfers electrons from low-molecular-weight aldehydes (e.g. glyoxal and glycoaldehyde) to O2 and so forms H2O2. Aryl alcohol oxidase is another H2O2-generating enzyme; in the process of converting benzyl alcohols to the aldehydes it transfers electrons to O2, generating H2O2. Ceriporiopsis subvermispora secretes an oxalate oxidase, which catalyses the degradation of oxalate to carbon dioxide and H2O2 and could be the main provider of H2O2 for manganese peroxidase in this organism.
Although few fungi produce ligninolytic enzymes, a much wider range excrete laccases as extracellular enzymes. These are copper-containing oxygenases which are able to oxidise o- and p-phenols and are required for the metabolism of lignin degradation products. They are particularly interesting as their appearance or disappearance in fungal cultures has been correlated with sexual and asexual reproduction in a number of cases. Thus, during mycelial growth of the cultivated mushroom, Agaricus bisporus, a large proportion of the compost lignin is degraded and correspondingly high activities of laccase are recorded (this one enzyme can amount to 2% of the total fungal protein). Yet, as the culture forms fruit bodies laccase activity is rapidly lost, initially by inactivation and subsequently by proteolysis; this pattern of behaviour reflects the changing nutritional demands of fungal mycelia as they process through successive developmental phases and the ability of the mycelium to act on its environment to satisfy those demands.
Laccases have been found in many fungi, including non-ligninolytic members of the Ascomycota, such as Aspergillus and Neurospora, as well as wood-rotting Basidiomycota. Laccases also occur in plants where they contribute to lignin biosynthesis. To react with a broader range of substrates laccases can interact with a mediator compound, which is a low-molecular-weight co-substrate secreted by the fungus that functions as a diffusible lignin-oxidising agent.
What's the relevance of this to organopollutants?
It is the ability of the fungi to degrade aromatic ring containing compounds such as lignin which make their enzymes applicable to organopollutants. As the enzymes are non-specific they are capable of degrading most aromatic complexes.
Lignin-degrading enzymes have considerable promise in several areas of biotechnology. Biopulping has already been mentioned as an industry where ligninolytic enzymes can improve the quality of pulp by releasing and purifying the cellulose. Laccases have potential, too, for:
- pulp bleaching;
- detoxification (particularly of pulp mill effluents);
- removal of phenolics from wines;
- chemical transformation of pharmaceuticals.
Further, many pesticidal treatments depend on benzene rings for their effectiveness; examples are chemicals like pentachlorophenol (PCP) and polychlorinated biphenyls (PCBs). These persist in the environment because there are so few organisms able to catabolise them. But organisms that can catabolise lignin have all the tools necessary to destroy such persistent pesticides (Fig. 1).
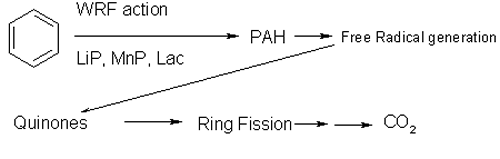 |
| Fig. 1. A simplified degradation pathway for polycyclic aromatic hydrocarbons by fungi. WRF = white rot fungi; LiP = lignin peroxidase; MnP = Manganese-dependent peroxidase; Lac = laccase; PAH = polycyclic aromatic hydrocarbon. |
Each lignin-degrading enzyme affects the organopollutants differently, and together they act on different parts to break down the entire substrate into carbon dioxide.
Laccase can catalyse both depolymerising reactions and also polymerising reactions. Whilst the depolymerising is obviously useful for the breakdown of the pollutants, polymerisation can also be useful, even though larger compounds are created. This is because sequestration is acceptable as a bioremedial method. Whilst forming a larger compound does not remove it from the environment, it can make the substance non-toxic, and thus negates the need for its removal.
Depolymerisation by laccase occurs by the oxidation of phenolic compounds in the pollutant which can result in an alkyl-phenyl cleavage, and consequently the aromatic ring is broken. The opened ring is a fatty acid which can relatively easily feed into normal metabolism.
The co-substrates produced assist due to their small size. This gives them the ability to access areas deep inside organopollutant matrices which are otherwise non-accessible to the enzymes themselves, thus improving the area available for degradation.
Manganese-dependent peroxidases produce a Mn³+ chelate oxalate which is small enough to diffuse into areas inaccessible to enzymes, which helps with organopollutants that are buried deep in soil which is not necessarily available to the fungal enzymes.
Lignin peroxidase appears to be the crucial enzyme for degradation of organopollutants, and some of the most successful experiments have used white rot fungi which produce lignin peroxidase. This is likely to be due to its ability to degrade substances with higher redox potentials.
Chloromethanes and bioluminescence
There are two intriguing side issues to fungal degradation of timber. One, which may become more crucial in the future, is that some of the small organic molecules the fungi produce to enhance activity of the lignin and manganese peroxidases incorporate chlorine from the wood into chloroaromatics, and even the synthesis of veratryl alcohol requires chloromethane. These volatile compounds are released into the atmosphere (thereby flushing the chlorine out of the substrate) and may themselves act as environmental pollutants. Some white-rot genera, particularly Phellinus and Inonotus, release enormous quantities of chloromethane into the atmosphere as they digest wood. The annual global release to the atmosphere from this source has been estimated at 160,000 tonnes, 75% of which is released from tropical and subtropical forests, with 86% being attributable to Phellinus spp. alone. Chloromethane is a powerful greenhouse gas and atmospheric pollutant which can have adverse effects on stratospheric ozone, yet in this case it is the product of a natural ecosystem.
The second intriguing aspect is that there are at least 64 species of fungi that are bioluminescent; they emit light from their mycelium and/or fruit bodies, and all species found so far have proved to be wood-decay fungi. All known truly bioluminescent fungi are white-spored agarics belonging to the traditional Family Trichomolataceae (Basidiomycota). So they are mushroom-forming, saprotrophic or, rarely, plant pathogenic species belonging to three distinct evolutionary lineages, which are named for their characteristic genera: 12 species belong to the Omphalotus lineage, 5 species to the Armillaria lineage (A. mellea is the most widely distributed bioluminescent fungus), and 47 species belong to the mycenoid lineage. Different luminous fungi emit light from different tissues, for example, from:
- mycelium and rhizomorphs in Armillaria;
- mycelium only in many Mycena species;
- spores only in Mycena rorida;
- sclerotia only of Collybia tuberosa;
- mycelium and the whole (mushroom) fruit body in Panellus stipticus and Omphalotus olearius (Clitocybe illudens); in the latter, light is emitted from the mushroom cap, stem, gills and spores.
Light emission in vitro can be obtained enzymatically by mixing cold extracts (which extract the enzymes) and hot extracts (which extract the substrates) from different species of fungi, which indicates a common mechanism for all these fungi. Kinetic data suggest a consecutive two-step enzymatic mechanism: first, a light-emitting substrate (arbitrarily called luciferin) is reduced by a soluble reductase at the expense of NAD(P)H; second, reduced luciferin is oxidised by an insoluble (membrane-bound?) luciferase that releases the energy in the form of bluish-green visible light with an emission maximum wavelength close to 530 nm.
Although fungal ‘luciferin’ is clearly one of the numerous secondary metabolites fungi can synthesise, its exact identity and structure has not yet been established. Nor is it entirely clear what the physiological and ecological function of fungal bioluminescence might be (which makes it difficult to appreciate its evolutionary selective advantage). It has been suggested that in the dark beneath closed tropical forest canopies bioluminescent fruit bodies may be at an advantage by attracting grazing animals (and that would include insects and other arthropods) that could help disperse their spores. Conversely, where mycelium (and vegetative structures like rhizomorphs and sclerotia) are the bioluminescent tissues, the argument has been made that light emission could deter grazing. Neither of these suggestions is entirely satisfactory. However, as far as is known at present, all luminescent basidiomycetes are white-rot fungi capable of lignin degradation. Bioluminescence is an oxygen-dependent metabolic process and may be used for detoxifcation of peroxides formed during ligninolysis, with the energy being released as light rather than heat. The favoured hypothesis at the moment, therefore, is that fungal bioluminescence is an advantageous process because it provides antioxidant protection against the potential deleterious effects of reactive oxygen species produced during wood decay.
Further information about ligninolytic enzymes can be found in the new textbook 21st Century Guidebook to Fungi by David Moore, Geoffrey D. Robson & Anthony P.J. Trinci. Published 2011 by Cambridge University Press: ISBN: 9780521186957. URL: ttp://www.cambridge.org/gb/knowledge/isbn/item6026594/?site_locale=en_GB. View Amazon page.
Updated December 15, 2016
